Archive
Logistic regression – simulation for a power calculation…
Please note – I’ve spotted a problem with the approach taken in this post – it seems to underestimate power in certain circumstances. I’ll post again with a correction or a more full explanation when I’ve sorted it.
So, I posted an answer on cross validation regarding logistic regression. I thought I’d post it in a little more depth here, with a few illustrative figures. It’s based on the approach which Stephen Kolassa described.
Power calculations for logistic regression are discussed in some detail in Hosmer and Lemeshow (Ch 8.5). One approach with R is to simulate a dataset a few thousand times, and see how often your dataset gets the p value right. If it does 95% of the time, then you have 95% power.
In this code we use the approach which Kleinman and Horton use to simulate data for a logistic regression. We then initially calculate the overall proportion of events. To change the number of events adjust odds.ratio. The independent variable is assumed to be normally distributed with mean 0 and variance 1.
nn <- 950
runs <- 10000
intercept <- log(9)
odds.ratio <- 1.5
beta <- log(odds.ratio)
proportion <- replicate(
n = runs,
expr = {
xtest <- rnorm(nn)
linpred <- intercept + (xtest * beta)
prob <- exp(linpred)/(1 + exp(linpred))
runis <- runif(length(xtest),0,1)
ytest <- ifelse(runis < prob,1,0)
prop <- length(which(ytest <= 0.5))/length(ytest)
}
)
summary(proportion)
This plot shows how the intercept and odds ratio affect the overall proportion of events per trial:
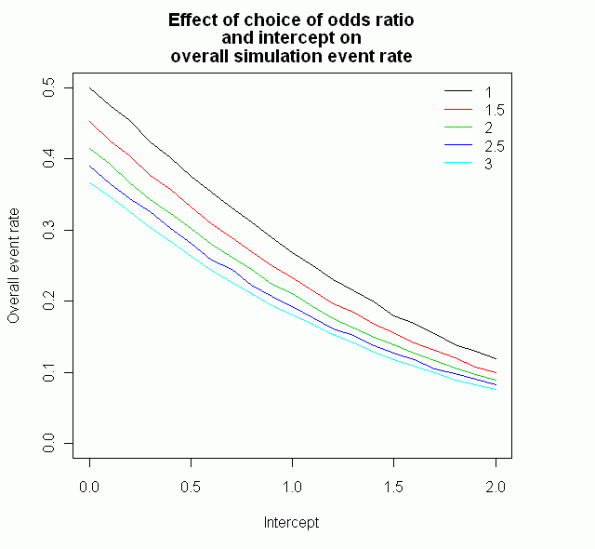
When you’re happy that the proportion of events is right (with some prior knowledge of the dataset), you can then fit a model and calculate a p value for that model. We use R’s inbuilt function replicate to do this 10,000 times, and count the proportion where it gets it right (i.e. p < 0.05). The proportion of the time that the simulation correctly get's the p < 0.05 is essentially the power of the logistic regression for your number of cases, odds ratio and intercept.
result <- replicate(
n = runs,
expr = {
xtest <- rnorm(nn)
linpred <- intercept + (xtest * beta)
prob <- exp(linpred)/(1 + exp(linpred))
runis <- runif(length(xtest),0,1)
ytest <- ifelse(runis < prob,1,0)
summary(model <- glm(ytest ~ xtest, family = "binomial"))$coefficients[2,4] < .05
}
)
print(sum(result)/runs)
I checked it against the examples given in Hsieh, 1999. It seemed to work pretty well calculating the power to be within ~ 1% of the power of the examples given in table II of that paper.
We can do some interesting things with R. I simulated a range of odds ratios and a range of sample sizes. The plot of these looks like this (each line represents an odd ratio):-
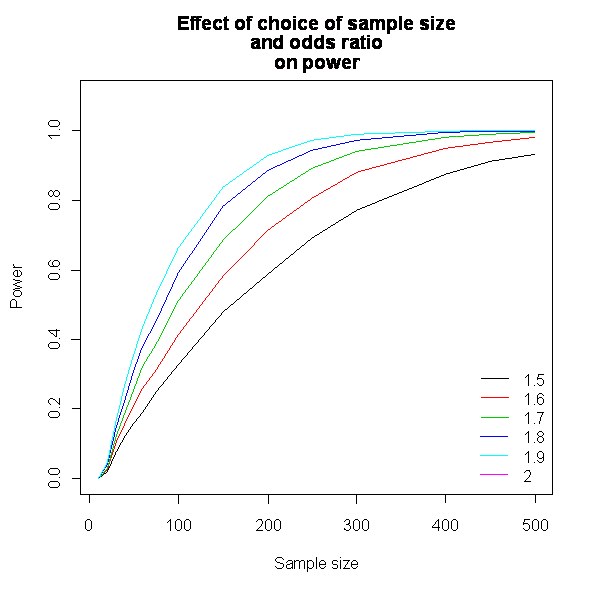
We can also keep the odds ratio constant, but adjust the proportion of events per trial. This looks like this (each line represents an event rate):
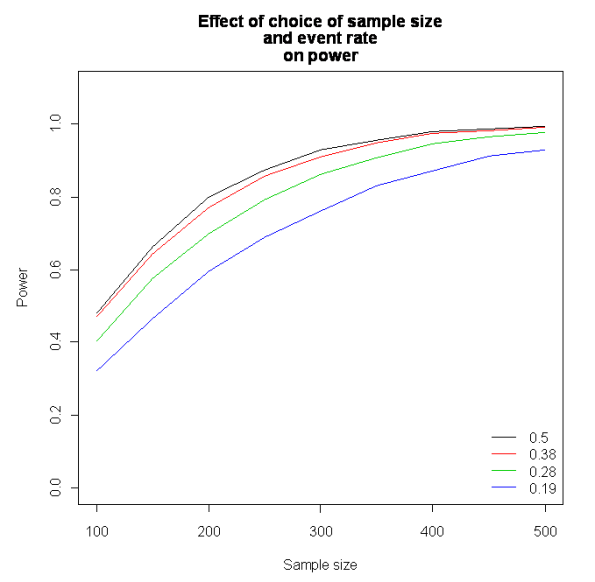
As ever, if anyone can spot an error or suggest a simpler way to do this then let me know. I haven’t tested my simulation against any packages which calculate power for logistic regression, but if anyone can it would be great to hear from you.
How to check if a file exists with HTTP and R
So, there’s probably an easier way to do this (please let me know if you know it)…
Suppose you’re working with a system which creates (binary) files and posts them for download on a website. You know the names of the files that will be created. However, they may not have been made yet (they’re generated on the fly, and appear in a vaguely random order over time). There are several of them and you want to know which ones are there yet, and when there are enough uploaded, run an analysis.
I spent quite a bit of time trying to work this out, and eventually came up with the following solution:
require(RCurl)
newurl <- c("http://cran.r-project.org/web/packages/RCurl/RCurl.pdf",
"http://cran.r-project.org/web/packages/RCurl/RCurl2.pdf")
for (n in 2:1){
z <- ""
try(z <- getBinaryURL(newurl[n], failonerror = TRUE))
if (length(z) > 1) {print(paste(newurl[n], " exists", sep = ""))
} else {print(paste(newurl[n], " doesn't exist", sep = ""))}
}
What this does is uses RCurl to download the file into a variable z. Then your system will check to see if z now contains the file.
If the file doesn’t exist, getBinaryURL() returns an error, and your loop (if you are doing several files) will quit. Wrapping the getBinaryURL() in try() means that the error won’t stop the loop from trying the next file (if you don’t trust me, try doing the above without the try wrapper). You can see how wrapping this in a loop could quickly go through several files and download ones which exist.
I’d really like to be able to do this, but not actually download the whole file (e.g. just the first 100 bytes) to see how many files of interest have been created, and if enough have, then download them all. I just can’t work out how to yet – I tried the range option of getBinaryURL() but this just crashed R. This would be useful if you are collecting data in real time, and you know you need at least (for example) 80% of the data to be available before you jump into a computationally expensive algorithm.
So, there must be an easier way to do all this, but can I find it? …